Publications
Refereed Journal Articles
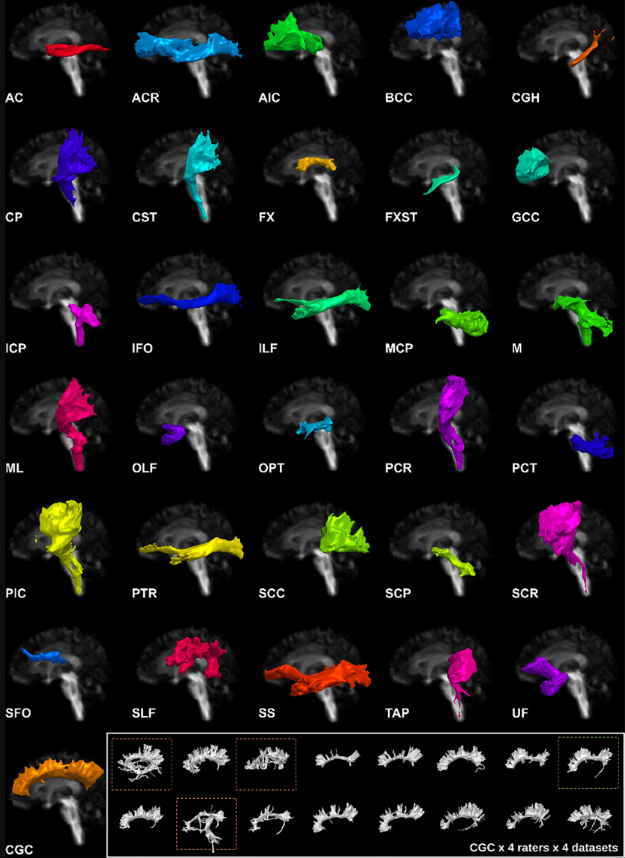
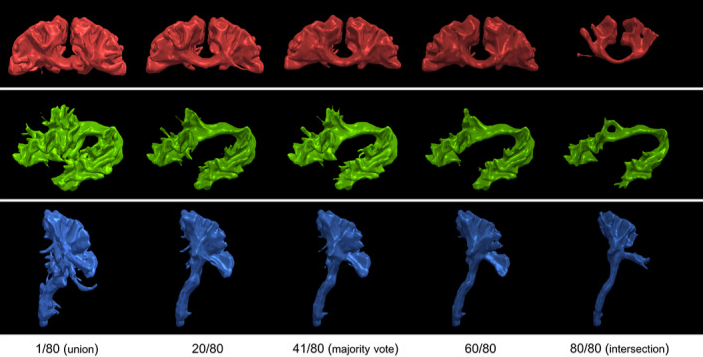
Efficient communication across fields of research is challenging, especially when they are at opposite ends of the physical and digital spectrum. Neuroanatomy and neuroimaging may seem close to each other. When neuroimaging studies try to isolate structures of interest, according to a specific anatomical definition, a variety of challenges emerge. It is a non-trivial task to convert the neuroanatomical knowledge to instructions and rules to be executed in neuroimaging software. In the process called “virtual dissection” used to isolate coherent white matter structure in tractography, each white matter pathway has its own set of landmarks (regions of interest) used as inclusion and exclusion criteria. The ability to segment and study these pathways is critical for scientific progress, yet, variability may depend on region placement, and be influenced by the person positioning the region (i.e., a rater). When raters’ variability is taken into account, the impact made by each region of interest becomes even more difficult to interpret. A delicate balance between anatomical validity, impact on the virtual dissection and raters’ reproducibility emerge. In this work, we investigate this balance by leveraging manual delineation data of a group of raters from a previous study to quantify which set of landmarks and criteria contribute most to variability in virtual dissection. To supplement our analysis, the variability of each pathway with a region-by-region exploration was performed. We present a detailed exploration and description of each region, the causes of variability and its impacts. Finally, we provide a brief overview of the lessons learned from our previous virtual dissection projects and propose recommendations for future virtual dissection protocols as well as perspectives to reach better community agreement when it comes to anatomical definitions of white matter pathways.
 Link
Link
Reproducible identification of white matter pathways across subjects is essential for the study of structural connectivity of the human brain. One of the key challenges is anatomical differences between subjects and human rater subjectivity in labeling. Labeling white matter regions of interest presents many challenges due to the need to integrate both local and global information. Clearly communicating the manual processes to capture this information is cumbersome, yet essential to lay a solid foundation for comprehensive atlases. Segmentation protocols must be designed so the interpretation of the requested tasks as well as locating structural landmarks is anatomically accurate, intuitive and reproducible. In this work, we quantified the reproducibility of a first iteration of an open/public multi-bundle segmentation protocol. This allowed us to establish a baseline for its reproducibility as well as to identify the limitations for future iterations. The protocol was tested/evaluated on both typical 3 T research acquisition Baltimore Longitudinal Study of Aging (BLSA) and high-acquisition quality Human Connectome Project (HCP) datasets. The results show that a rudimentary protocol can produce acceptable intra-rater and inter-rater reproducibility. However, this work highlights the difficulty in generalizing reproducible results and the importance of reaching consensus on anatomical description of white matter pathways. The protocol has been made available in open source to improve generalizability and reliability in collaboration. The goal is to improve upon the first iteration and initiate a discussion on the anatomical validity (or lack thereof) of some bundle definitions and the importance of reproducibility of tractography segmentation.
 Link
Link
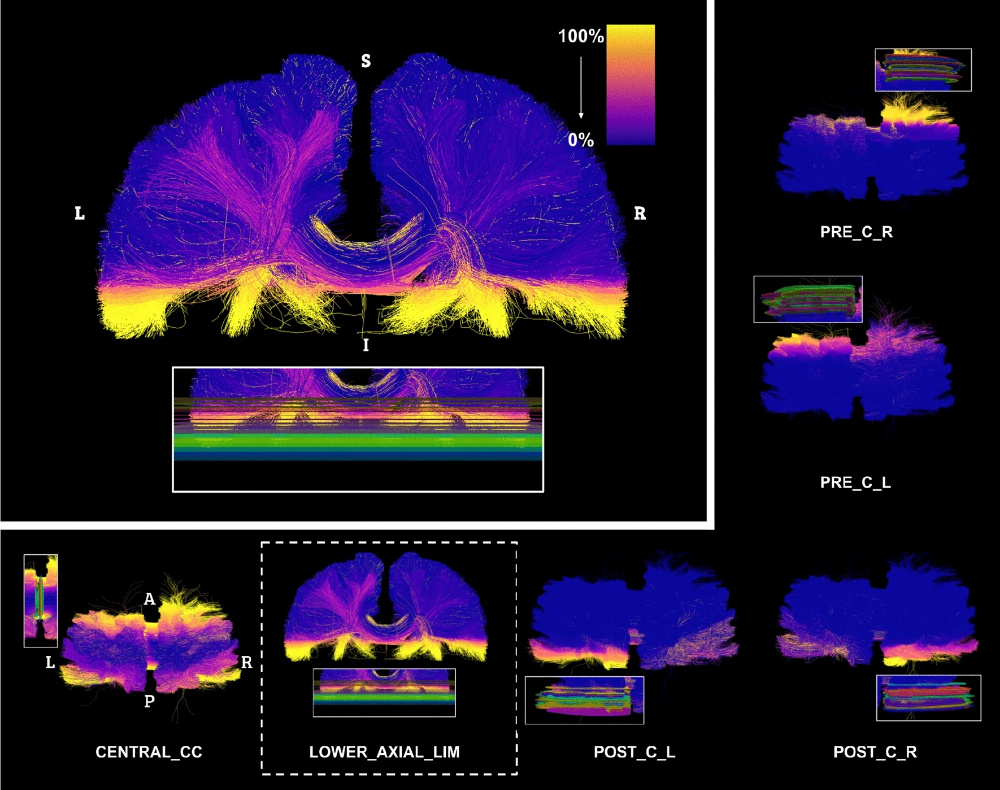
The segmentation of brain structures is a key component of many neuroimaging studies. Consistent anatomical definitions are crucial to ensure consensus on the position and shape of brain structures, but segmentations are prone to variation in their interpretation and execution. White-matter (WM) pathways are global structures of the brain defined by local landmarks, which leads to anatomical definitions being difficult to convey, learn, or teach. Moreover, the complex shape of WM pathways and their representation using tractography (streamlines) make the design and evaluation of dissection protocols difficult and time-consuming. The first iteration of Tractostorm quantified the variability of a pyramidal tract dissection protocol and compared results between experts in neuroanatomy and nonexperts. Despite virtual dissection being used for decades, in-depth investigations of how learning or practicing such protocols impact dissection results are nonexistent. To begin to fill the gap, we evaluate an online educational tractography course and investigate the impact learning and practicing a dissection protocol has on interrater (groupwise) reproducibility. To generate the required data to quantify reproducibility across raters and time, 20 independent raters performed dissections of three bundles of interest on five Human Connectome Project subjects, each with four timepoints. Our investigation shows that the dissection protocol in conjunction with an online course achieves a high level of reproducibility (between 0.85 and 0.90 for the voxel-based Dice score) for the three bundles of interest and remains stable over time (repetition of the protocol). Suggesting that once raters are familiar with the software and tasks at hand, their interpretation and execution at the group level do not drastically vary. When compared to previous work that used a different method of communication for the protocol, our results show that incorporating a virtual educational session increased reproducibility. Insights from this work may be used to improve the future design of WM pathway dissection protocols and to further inform neuroanatomical definitions.
 Link
Link
The human brain is a complex and organized network, where the connection between regions is not achieved with single axons crisscrossing each other but rather millions of densely packed and well-ordered axons. Reconstruction from diffusion MRI tractography is only an attempt to capture the full complexity of this network, at the macroscale. This review provides an overview of the misconceptions, biases and pitfalls present in structural white matter bundle and connectome reconstruction using tractography. The goal is not to discourage readers, but rather to inform them of the limitations present in the methods used by researchers in the field in order to focus on what they can do and promote proper interpretations of their results. It also provides a list of open problems that could be solved in future research projects for the next generation of PhD students.
 Link
Link
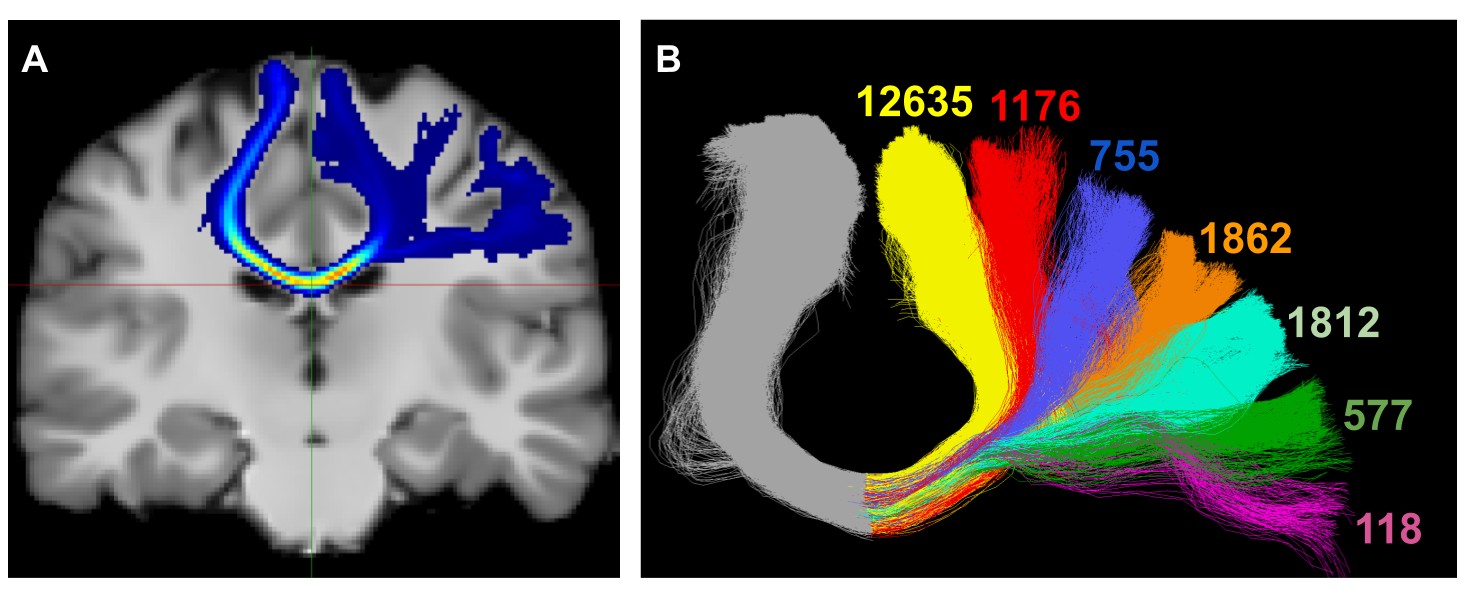
Investigative studies of white matter (WM) brain structures using diffusion MRI (dMRI) tractography frequently require manual WM bundle segmentation, often called “virtual dissection.” Human errors and personal decisions make these manual segmentations hard to reproduce, which have not yet been quantified by the dMRI community. It is our opinion that if the field of dMRI tractography wants to be taken seriously as a widespread clinical tool, it is imperative to harmonize WM bundle segmentations and develop protocols aimed to be used in clinical settings. The EADC-ADNI Harmonized Hippocampal Protocol achieved such standardization through a series of steps that must be reproduced for every WM bundle. This article is an observation of the problematic. A specific bundle segmentation protocol was used in order to provide a real-life example, but the contribution of this article is to discuss the need for reproducibility and standardized protocol, as for any measurement tool. This study required the participation of 11 experts and 13 nonexperts in neuroanatomy and “virtual dissection” across various laboratories and hospitals. Intra-rater agreement (Dice score) was approximately 0.77, while inter-rater was approximately 0.65. The protocol provided to participants was not necessarily optimal, but its design mimics, in essence, what will be required in future protocols. Reporting tractometry results such as average fractional anisotropy, volume or streamline count of a particular bundle without a sufficient reproducibility score could make the analysis and interpretations more difficult. Coordinated efforts by the diffusion MRI tractography community are needed to quantify and account for reproducibility of WM bundle extraction protocols in this era of open and collaborative science.
 Link
Link
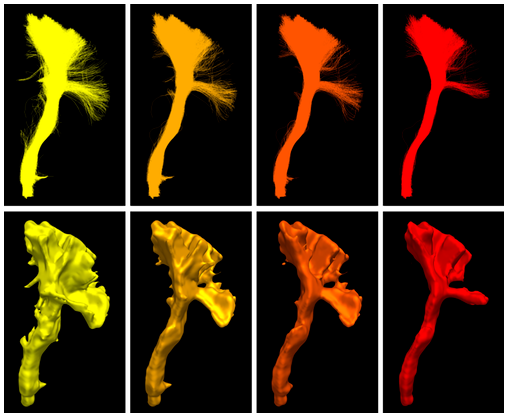
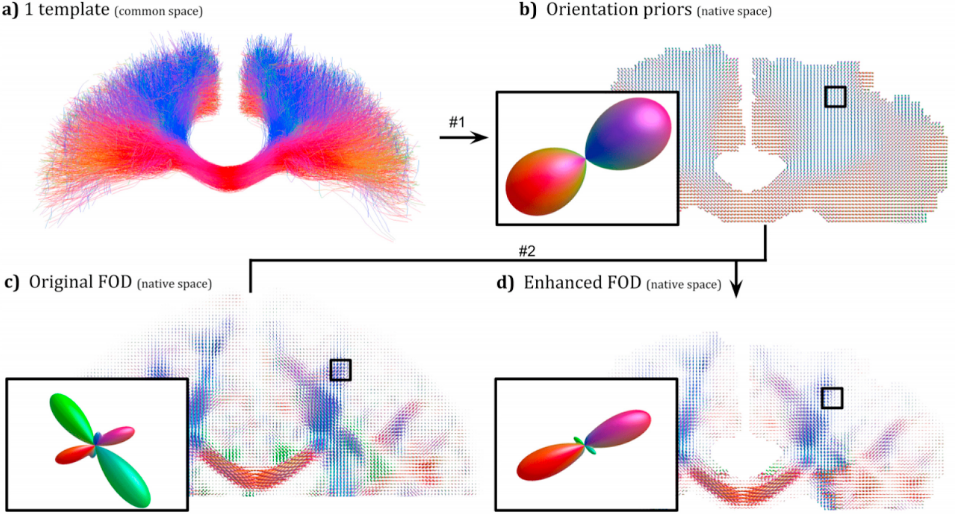
Anatomical white matter bundles vary in shape, size, length, and complexity, making diffusion MRI tractography reconstruction of some bundles more difficult than others. As a result, bundles reconstruction often suffers from a poor spatial extent recovery. To fill-up the white matter volume as much and as best as possible, millions of streamlines can be generated and filtering techniques applied to address this issue. However, well-known problems and biases are introduced such as the creation of a large number of false positives and over-representation of easy-to-track parts of bundles and under-representation of hard-to-track. To address these challenges, we developed a Bundle-Specific Tractography (BST) algorithm. It incorporates anatomical and orientational prior knowledge during the process of streamline tracing to increase reproducibility, sensitivity, specificity and efficiency when reconstructing certain bundles of interest. BST outperforms classical deterministic, probabilistic, and global tractography methods. The increase in anatomically plausible streamlines, with larger spatial coverage, helps to accurately represent the full shape of bundles, which could greatly enhance and robustify tract-based and connectivity-based neuroimaging studies.
 Link
Link
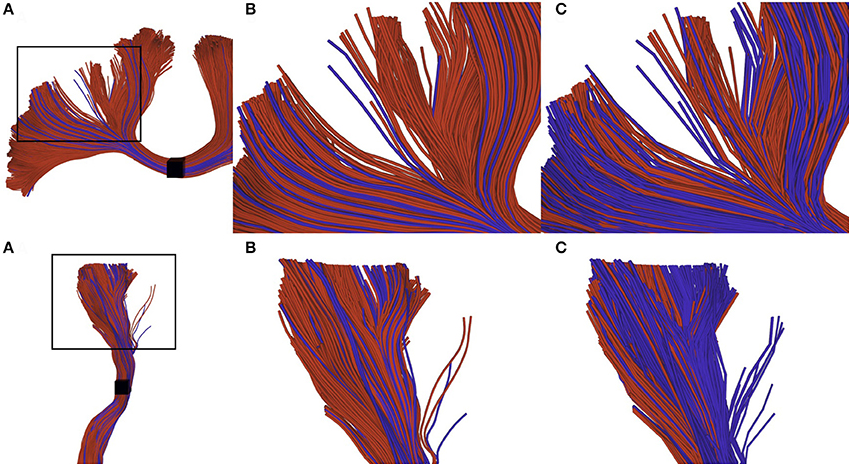
Recently proposed tractography and connectomics approaches often require a very large number of streamlines, in the order of millions. Generating, storing and interacting with these datasets is currently quite difficult, since they require a lot of space in memory and processing time. Compression is a common approach to reduce data size. Recently such an approach has been proposed consisting in removing collinear points in the streamlines. Removing points from streamlines results in files that cannot be robustly post-processed and interacted with existing tools, which are for the most part point-based. The aim of this work is to improve visualization, interaction and tractometry algorithms to robustly handle compressed tractography datasets. Our proposed improvements are threefold: (i) An efficient loading procedure to improve visualization (reduce memory usage up to 95% for a 0.2 mm step size); (ii) interaction techniques robust to compressed tractograms; (iii) tractometry techniques robust to compressed tractograms to eliminate biased in tract-based statistics. The present work demonstrates the need of correctly handling compressed streamlines to avoid biases in future tractometry and connectomics studies.
 Link
Link
Preprints, Books (chapter), and Technical Reports
Tractography is known to have problems reconstructing white matter bundles that are narrow, have high curvature, or go through partial volume voxels contaminated by CSF or gray matter. One such bundle is the fornix, the major output tract of the hippocampus, which is especially problematic with aging. Hippocampal atrophy and ventricular expansion make the fornix even harder (often impossible) to track with current state-of-the-art techniques. In this work, a bundle-specific tractography algorithm is proposed to fully reconstruct the fornix. By injecting shape, position, and orientation priors, fornix reconstruction is markedly is improved. We report an increase in spatial coverage and better reproducibility across test-retest. These improvements over classical tractography algorithms also enable tractometry of the fornix to be combined with dual-tracer positron emission tomography (PET) data in participants with mild cognitive impairment (MCI). MCI participants underwent a multi-modal brain imaging before and after a 6-month daily ketogenic supplement. We report, for the first time, significant diffusion measures and 18F-fluorodeoxyglucose (FDG) uptake differences in specific sub-sections of the fornix after the ketogenic supplement.
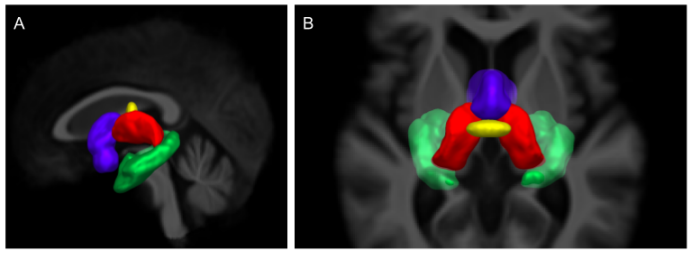 Link
Link
Conference Papers
Tractography allows the investigation of white matter fascicles. However, it requires a large amount of streamlines to be generated to cover the full spatial extent of desired bundles. In this work, a bundle-specific tractography algorithm was developed to increase reproducibility and sensitivity of white matter fascicle virtual dissection, thus avoiding the computation of a full brain tractography. Using fascicle priors from manually segmented bundles templates or atlases, we propose a novel local orientation enhancement methodology that overcomes reconstruction difficulties in crossing regions. To reduce unnecessary computation, tractography seeding and tracking were restricted to specific locales within the brain. These additions yield better spatial coverage, increasing the quality of the fanning in crossing regions, helping to accurately represent fascicle shape. In this work, tractography methods were analyzed and compared using a single bundle of interest, the corticospinal tract.
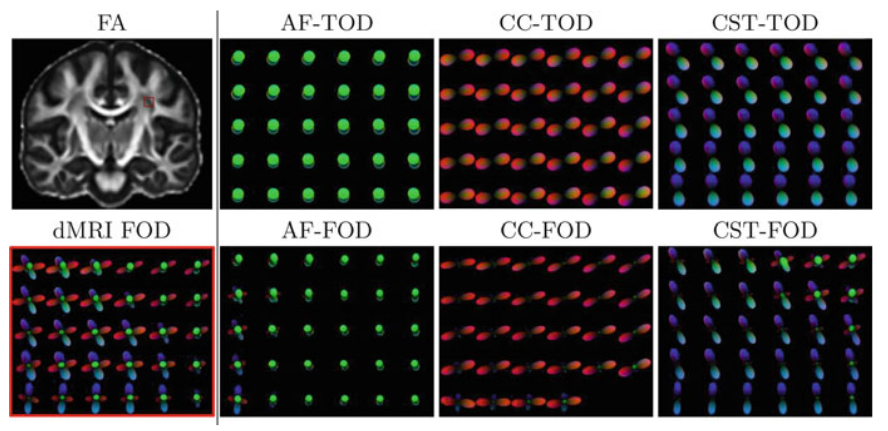 Link
Link
Theses and memoirs
L'imagerie par résonance magnétique de diffusion (IRMd) est une modalité d'acquisition permettant de sonder les tissus biologiques et d'en extraire une variété d'informations sur le mouvement microscopique des molécules d'eau. Plus spécifiquement à l'imagerie médicale, l'IRMd permet l'investigation des structures fibreuses de nombreux organes et facilite la compréhension des processus cognitifs ou au diagnostic. Dans le domaine des neurosciences, l'IRMd est cruciale à l'exploration de la connectivité structurelle de la matière blanche. Cette thèse s'intéresse plus particulièrement à la reconstruction de faisceaux de la matière blanche ainsi qu'à leur analyse. Toute la complexité du traitement des données commençant au scanneur jusqu'à la création d'un tractogramme est extrêmement importante. Par contre, l'application spécifique de reconstruction des faisceaux anatomiques plausibles est ultimement le véritable défi de l'IRMd. L'optimisation des paramètres de la tractographie, le processus de segmentation manuelle ou automatique ainsi que l'interprétation des résultats liée à ces faisceaux sont toutes des étapes du processus avec leurs lots de difficultés.
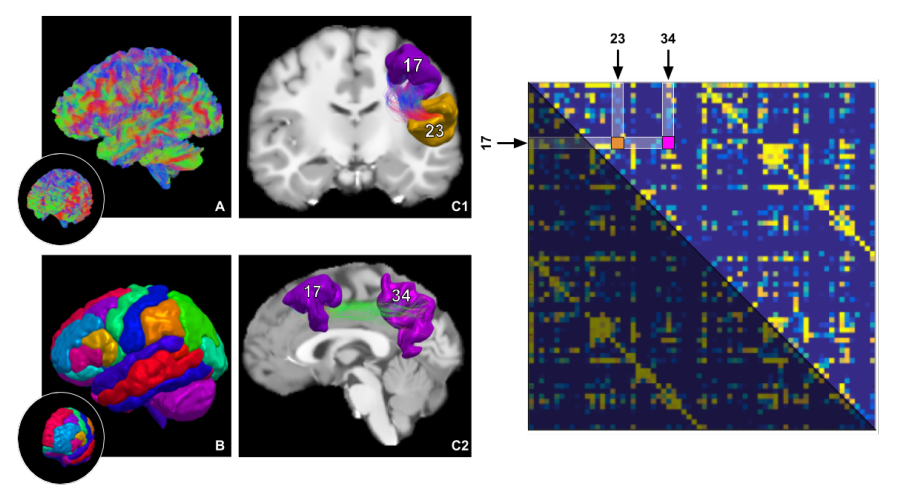 Link
Link